use cases

In this repo, we present a full example of the metaGOflow pipeline using actual EMO BON sequence data.
metaGOflow is a pipeline based on MGnify and its workflows, aiming to address the challenges of the analysis of the European Marine Omics Biodiversity Observation Network (EMO BON) data. EMO BON is a long-term omics observatory of marine biodiversity that generates hundreds of metagenomic samples periodically from a range of stations around Europe.
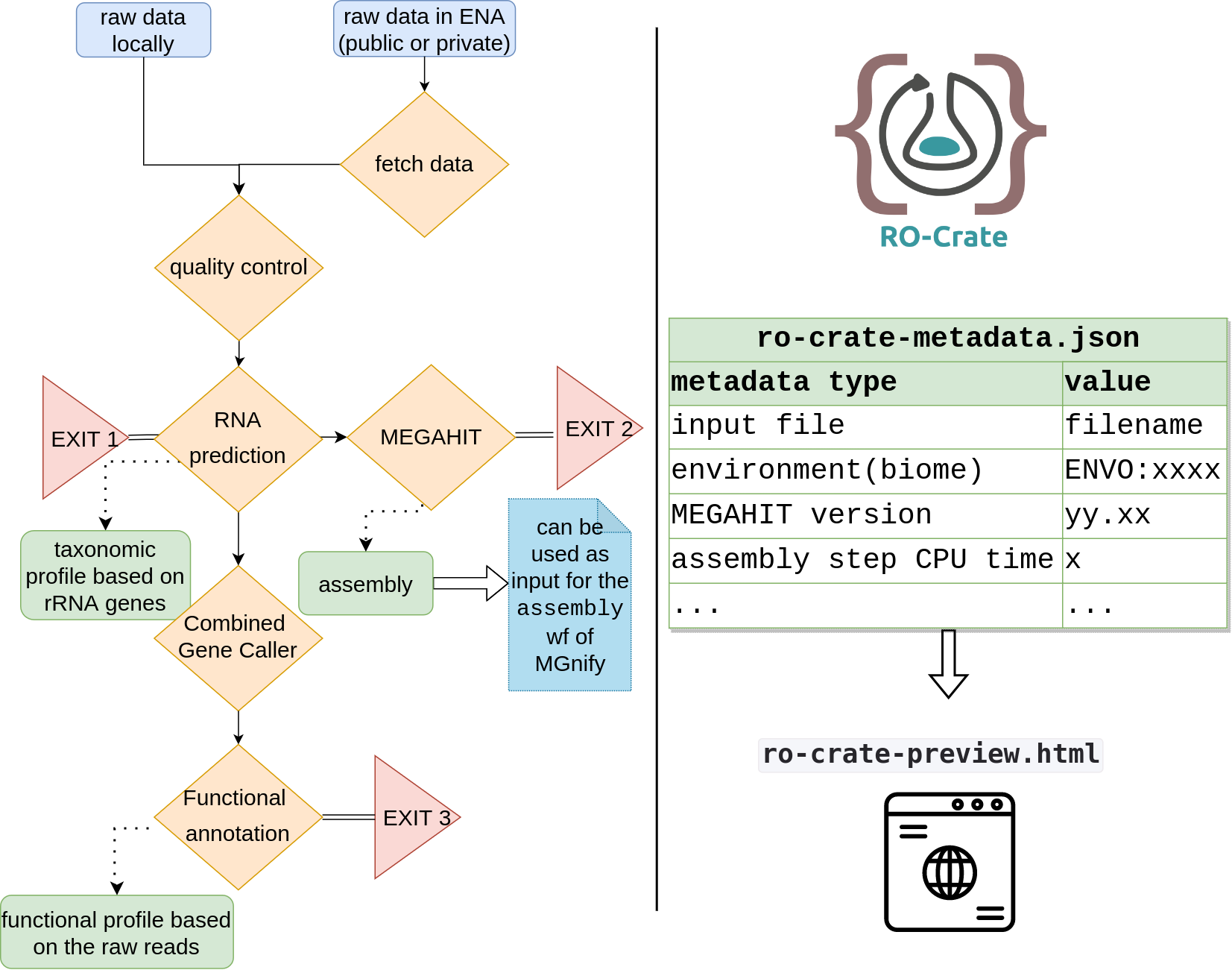
In the following diagram is an overview of the metaGOflow steps:
Here we show visual components accompanying the metaGOflow publication. Along with the EMO BON samples included in this use case, a TARA OCEAN sample of size similar to the EMO BON data was analysed. A quality control report, the taxonomic inventories as well as some of the functional annotations returned in each case are displayed here. metaGOflow also returned the assemblies of the samples. You can find all the input-output files along with the RO-crates produced by metaGOflow for those 2 samples under this Zenodo repository.
-
Here are parts of the metaGOflow data products for the EMO BON marine sediment.
-
Here are again parts of the metaGOflow data products for the EMO BON water column sample.
-
Last, here is an overview on the metaGOflow data products when using the TARA OCEANS sample
metaGOflow source code: GitHub repo
Citation
In case you are using metaGOflow for your analysis, please remember to cite us:
Contact
For bugs/issues you may open an issue on the GitHub repository of metaGOflow or email Dr. Haris Zafeiropoulos (haris.zafeiropoulos@kuleuven.be)
With respect to EMO BON protocols, samples, analyses you may contact the Observation, Data and Service Development Officer of EMBRC, Dr. Ioulia Santi (ioulia.santi@embrc.eu)
![]()