Microbiome basics

Microbiome analysis counts several aspects, yet it always comes down at some basic concepts! Here, I will try to cover a thing or two and gather some links that describe them more thoroughly. To save some (of my) time, I will use a lot of copy-paste () always referring the initial resource
TABLE OF CONTENTS
- Microbiome data idiosyncrasy
- Microbiome diversity
- Feature Identification
- Microbial phenomena that can drive you nuts!
- Literature
Microbiome data idiosyncrasy
Although microbiome data has some of the attributes of compositional data, it is not perfectly compositional. Classic compositional data vectors represent portions of a whole. The total sum of the components is not meaningful, and only the relative difference between components matters [36]. For truly compositional data, the vectors (2, 1) and (2000, 1000) represent the same information: only that the first and second components are present in the ratio 2 : 1. For microbiome data, the size of the counts also contains information about the reliability of the ratio. Larger counts are more likely to closely match the true ratio in the sample [44].
Covariate Adjustment
Regression analysis, potentially with penalization for variable selection, has been used to analyze an outcome of interest modeled as a function of microbiome features.
Microbiome diversity
Diversity within a community
α-diversity
https://docs.onecodex.com/en/articles/4136553-alpha-diversity
Shannon Entropy ((H’)) \[H' = - \sum_{i=1}^S p_i \ln(p_i)\]
where:
- \(p_i\) is the proportional abundance of species (i),
- \(S\) is the total number of species.
The effective number of species based on Shannon entropy is: \[\text{Effective Number of Species} = e^{H'}\]
Diversity between communities
β-diversity
Feature Identification
This sections is paraphrasing in a summary of the Liu et al. (2021) chapter in the Statistical Analysis of Microbiome Data book.
Questions to be addressed:
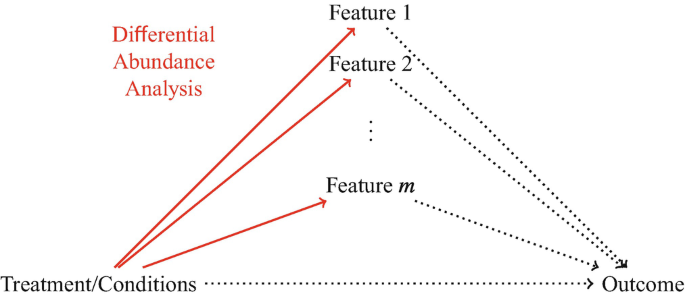
- which microbiome features are impacted by treatments or environmental conditions? \(\rightarrow\) identify features whose abundances change across treatments or conditions differential \(\rightarrow\) abundance analysis
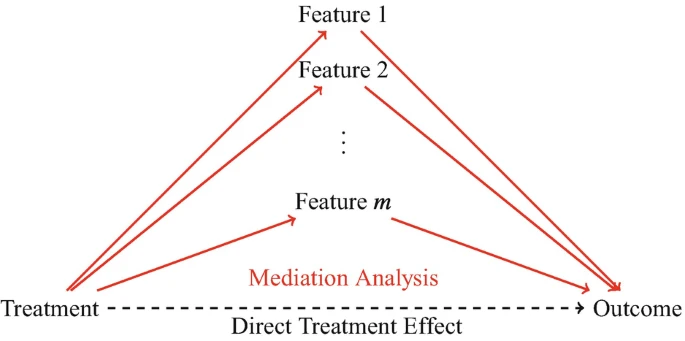
- which microbiome features mediate treatment effects on an outcome? \(\rightarrow\) identify taxa affected by treatments and that because of their change the outcome of the treatment is influenced \(\rightarrow\) mediation analysis
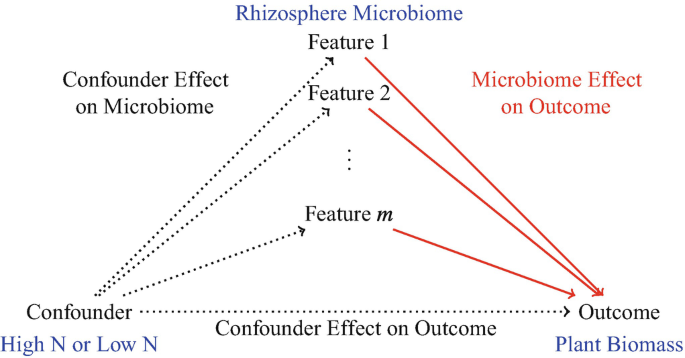
- which microbiome features have an effect on an outcome, adjusting for confounders \(\rightarrow\) identify microbiome features with an effect on an outcome with no particular treatments of interest, but with potential complex confounding arising from relationships between microbes, host, and environment \(\rightarrow\) Feature Identification Adjusting for Confounding
Differential Abundance Analysis
the null hypothesis of a differential abundance test is that treatments do not affect the mean abundance level.
Differential abundance analysis (solid lines) selects microbiome features whose abundance levels change across treatments or conditions. It only examines the relationship between treatments/conditions and microbiome features, but not the relationships involving other outcomes
Mediation analysis
high dimensionality and sparsity of microbiome data.
In case of a treatment - microbiome - outcome study, one needs to avoid differential abundance analysis as the relationships you are looking for are out of scope for DA!
Mediation analysis examines the indirect effects of treatment on the outcome through the microbiome. To determine whether a feature has a mediation effect, a method must consider both the effect of the treatment on the feature and the effect of the feature on the outcome.
Feature Identification Adjusting for Confounding
asd
Microbial phenomena that can drive you nuts!
A strain dips to very low abundance in a microbial community and then recovers to thrive
This is quite common and reflects the complex interplay of ecological, evolutionary, and environmental factors. In this page you can find some of the ways a strain might manage to survive during these low-abundance periods without going completely extinct.
###
Literature
Datta, Somnath, and Subharup Guha, eds. Statistical Analysis of Microbiome Data. Springer International Publishing, 2021.
