dingo

dingo is a Python package that supports a variety of methods to sample from the flux space of metabolic models, based on state-of-the-art random walks and rounding methods. It relies on high dimensional sampling with Markov Chain Monte Carlo (MCMC) methods and fast optimization methods to analyze the possible states of a metabolic network. To perform MCMC sampling, dingo relies on the C++ library volesti, which provides several algorithms for sampling convex polytopes. Among the different ways to sample, dingo also implements the Multiphase Monte Carlo Sampling algorithm (see post for relative publication).
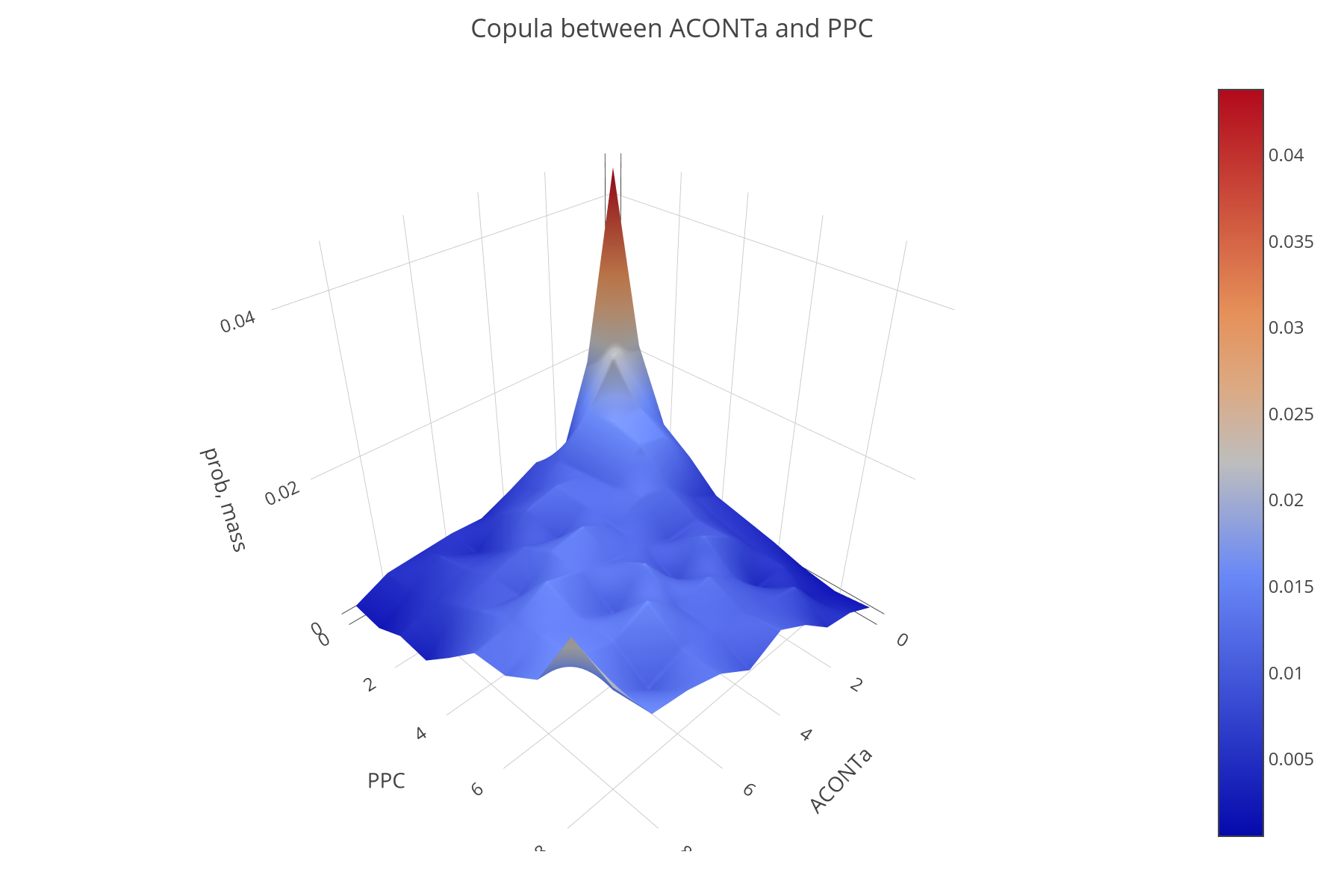
Flux sampling provides insgith of strong statistical evidence. For example, pairwise fluxes correlated with one another in a positive or negative way, can be found.
dingo also supports Flux Balance Analysis and Flux Variability Analysis, two standard methods to analyze the flux space of a metabolic network,.
dingo is part of the GeomScale that is over the last year has been an organization of Google Summer of Code.
