annotating microbial co-occurrence networks
background, documentation and a use case
CytoscapeApp View it on GitHub Join us on Matrix
About
Microbial interactions play a fundamental role in deciphering the underlying mechanisms that govern ecosystem functioning. Co-occurrence networks have been widely used for inferring microbial associations or/and interactions from metagenomic data. However, spurious associations and tool - dependence confine the network inference. The integration of previous evidence or/and knowledge can increase or decrease the confidence level of the retrieved associations. This way, associations can be further investigated, and more reliable conclusions can be drawn.
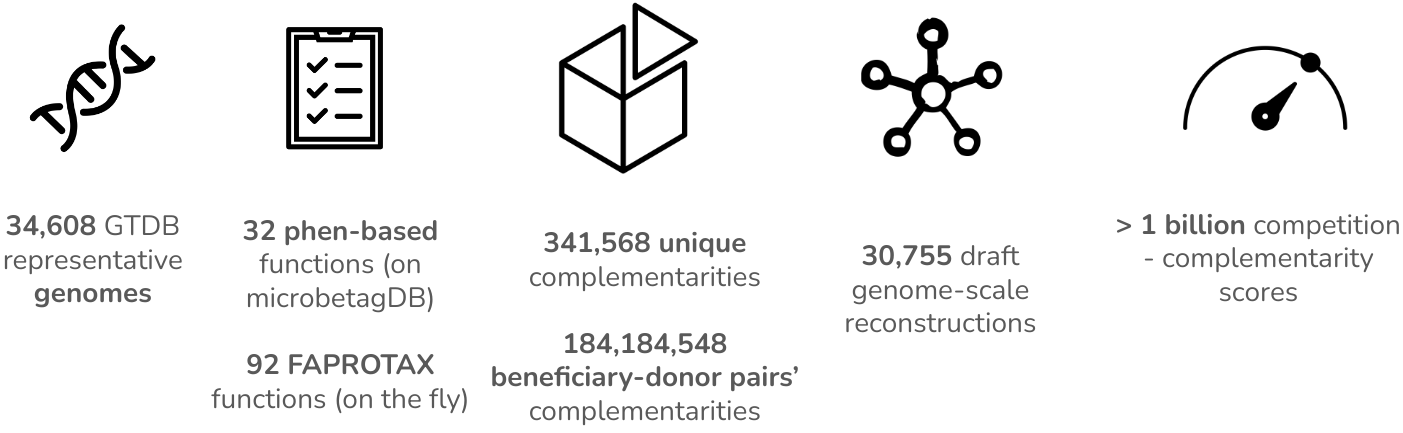
microbetag implements data integration techniques to annotate both the nodes (taxa) and the edges (predicted associations) of such a network to enhance microbial co-occurrence network analysis for amplicon data. Have a look at the modules tab to get an overview of the methods used.
How to use
microbetag is a software ecosystem with different software packages to use based on the tasks you are going for.
The most common use is through its graphical interface, a Cytoscape app called MGG. Cytoscape is a well-established, widely used software for network data integration, analysis, and visualization.
To use MGG you need to first make sure you have Cytoscape installed on your machine; if not you can do this from the Cytoscape Install page Then, there are two ways to install MGG in Cytoscape: Either from through the Cytoscape app store on a browser or from within Cytoscape. In the first case, you need to first lunching Cytoscape, and then visit the MGG Cytoscape Appstore page. By clicking the Install button MGG will be automatically added on your Cytoscape. Alternatively, to install MGG from within Cytoscape, you may click Apps > App Store > Show App Store, then search for “microbetag” in the pop-up box and follow this will guide you to the MGG page.
Once MGG is installed, you are ready to use microbetag either on the fly, by providing an OTUs/ASVs (amplicon data) and optionally a network, if you already have one, or locally, if you want to apply the annotations on your own bins/MAGs. In the last case, you will also have to install Docker or Singularity and pull the microbetag image that allows you to do so (see Additional tutorials for more).
HOW TO USE AND INTERPRET MICROBETAG’s FINDINGS
For a thorough description of the app, please check the Cytoscape App tab.
In addition, microbetag’s annotations are also available through its Application Programming Interface (API). This way, one may have direct access to the microbetagDB and may export annotations for species or pairs of species of interest, without the need of a network.
Contact
For hints on how to use microbetag, ideas for new features and bug reports find us on out Matrix space. If you do not have a Matrix account, it’s only two clicks away! For more information, you may check here.
Cite us
In prep.
Funding
This project was funded by an EMBO Scientific Exchange Grants and the 3D’omics Horizon 2020 project (101000309).
License
microbetag is under GNU General Public License v3.0. For third-party components separate licenses apply. The MGG CytoscapeApp is under Apache License, Version 2.0.