Run microbetag Cytoscape app
microbetag v1.0.1
MGG v1.0.0
microbetag web-app MGG source code Tutorial files
INPUT FILES AND PARAMETERS SETTING
Please, download the files required for the different cases and set the parameters as shown in this tutorial. Parameters are essential especially for the online version of
microbetagas they can lead either to non-optimal annotations or even failures of the software. You may check the FAQs section for rules of thumb on how to set your parameters and you are always welcome to join us on Matrix and ask us directly.
We show how to install and use the microbetag Cytoscape app (called MGG) and use it with your data to get microbetag-annotated networks using the micrbetagDB and the online version of microbetag. We also highlight the MGG features that allow you to go through the nodes and the edges annotations returned.
To start using microbetag you need first, to make sure you have Cytoscape on your system; if not, go ahead and download Cytoscape. Then, you need to install the microbetag app (MGG) from Cytoscape App store. Make sure you first lunch Cytoscape and then visit Cytoscape Appstore. If you have already visited the MGG page on Cytoscape Appstore, lunch Cytoscape and refresh the Cytoscape Appstore page. You should now see an Install button.
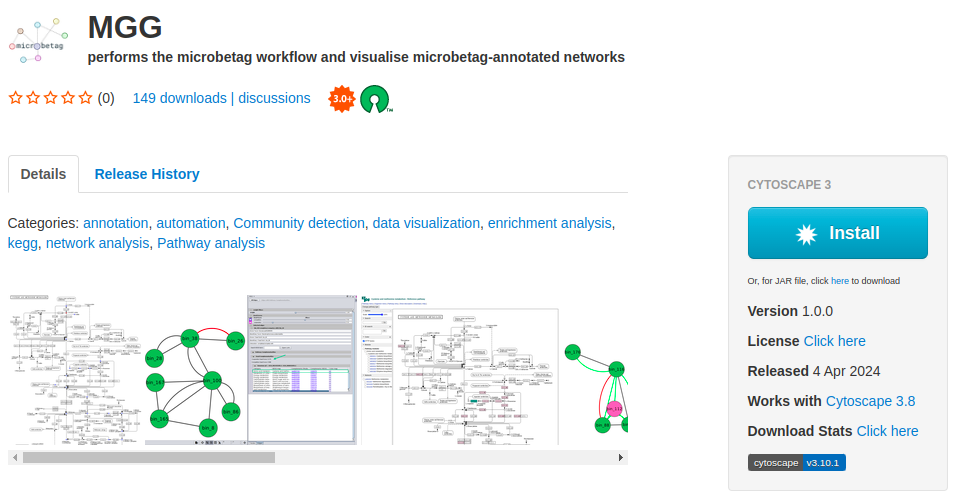
By clicking it, it will be automatically integrated on your Cytoscape. If you visit Cytoscape Appstore and you have not lunched Cytoscape, you will see a Download button instead of the Install. As already mentioned, we suggest you lunch Cytoscape and refresh the page. Otherwise, you can click the Download button and move manually the .jar file to the apps folder of your Cytoscape.
You can also get MGG from within Cytoscape by clicking on the Apps tab of the main bar and then App Store > Show App Store and typing microbetag on the box that pops up.
Once the app is installed, you may click on the Apps tab, and you will find MGG there.
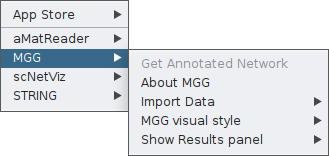
From the main menu box, you will have access to all features of the app. As you see, the Get Annotated Network is currently not a clickable option. That is because microbetag has no input yet.
You need first to feed the app with your abundance table and, if available, your co-occurrence network. In both cases though, the abundance table will be required.
Please, make sure your taxonomy fits the criteria for microbetag to run. You may find more on that issue on the Input files section.
Then, as you will see in the following two cases, you will have to set the values to a set of parameters to describe your input data but also what annotation steps you would like microbetag to perform.
| Parameter | Variable | Description | Value |
|---|---|---|---|
| Choose input type | input_category | In case you already have a network, set it as network and load it; otherwise set it as abundance_table. In both cases you need to provide the abundance table though | [abundance_table | network] |
| Choose taxonomy database | taxonomy | In case a user’s taxonomy is to be used, denotes which taxonomy scheme to be used from microbetag | [GTDB | dada2 | qiime2] |
| phenDB annotations | phenDB | return phenotypic traits based on phen models | bool |
| FAPROTAX annotations | faprotax | return annotations using the FAPROTAX database | bool |
| Pathway Complementarity | pathway_complement | return pathway complementarities between associated nodes | bool |
| Seed scores and complements | seed_scores | return complementarity and cooperation scores based on metabolic reconstructions seed sets | bool |
| Network clustering | manta | return clusters of nodes on the network using the manta package | bool |
| Consider children taxa | get_children | use genomes of children taxa of the taxa in the abundance table based on the NCBI Taxonomy scheme, relevant only if you use Other taxonomy | bool |
| heterogeneous | heterogeneous | (FlashWeave) enable heterogeneous mode for multi-habitat or -protocol data with at least thousands of samples (FlashWeaveHE) | bool |
| sensitive | sensitive | (FlashWeave) enable fine-grained associations (FlashWeave-S, FlashWeaveHE-S), sensitive=false results in the fast modes FlashWeave-F or FlashWeaveHE-F | bool |
The column Variables in the above table provides the variable names you need to use in case you are about to use microbetag from Python (see tutorial).
The datasets to be used in all cases except of the Using a network tutorial, are subsets of abundance tables with no special biological meaning. However, in the Using a network case, we do use the network of Hessler et al. (2023) who we would like to thank for sharing their data.